| Webtool | Description |
|---|
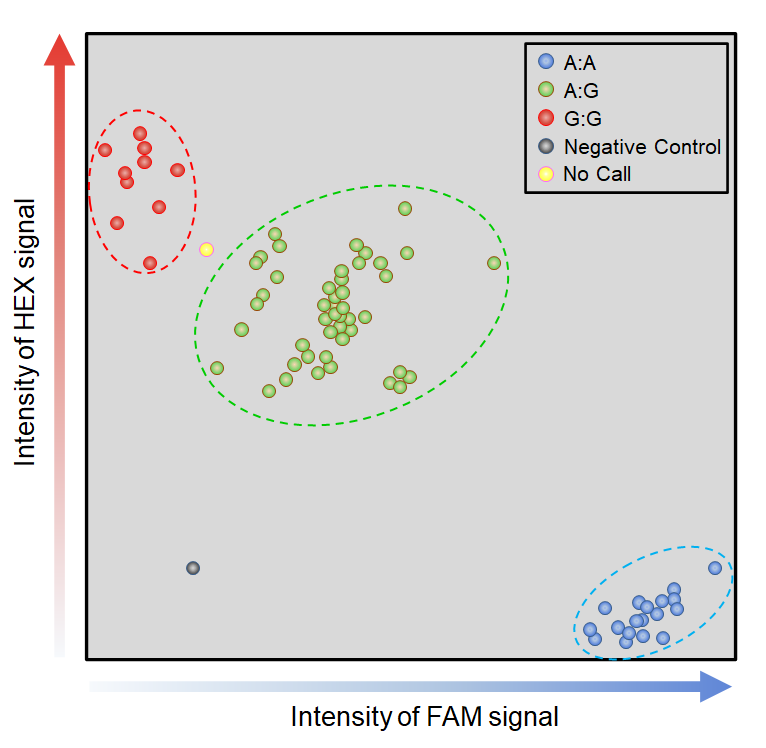 | Retrieve genotyping data derived from the 820K and 35K arrays. |
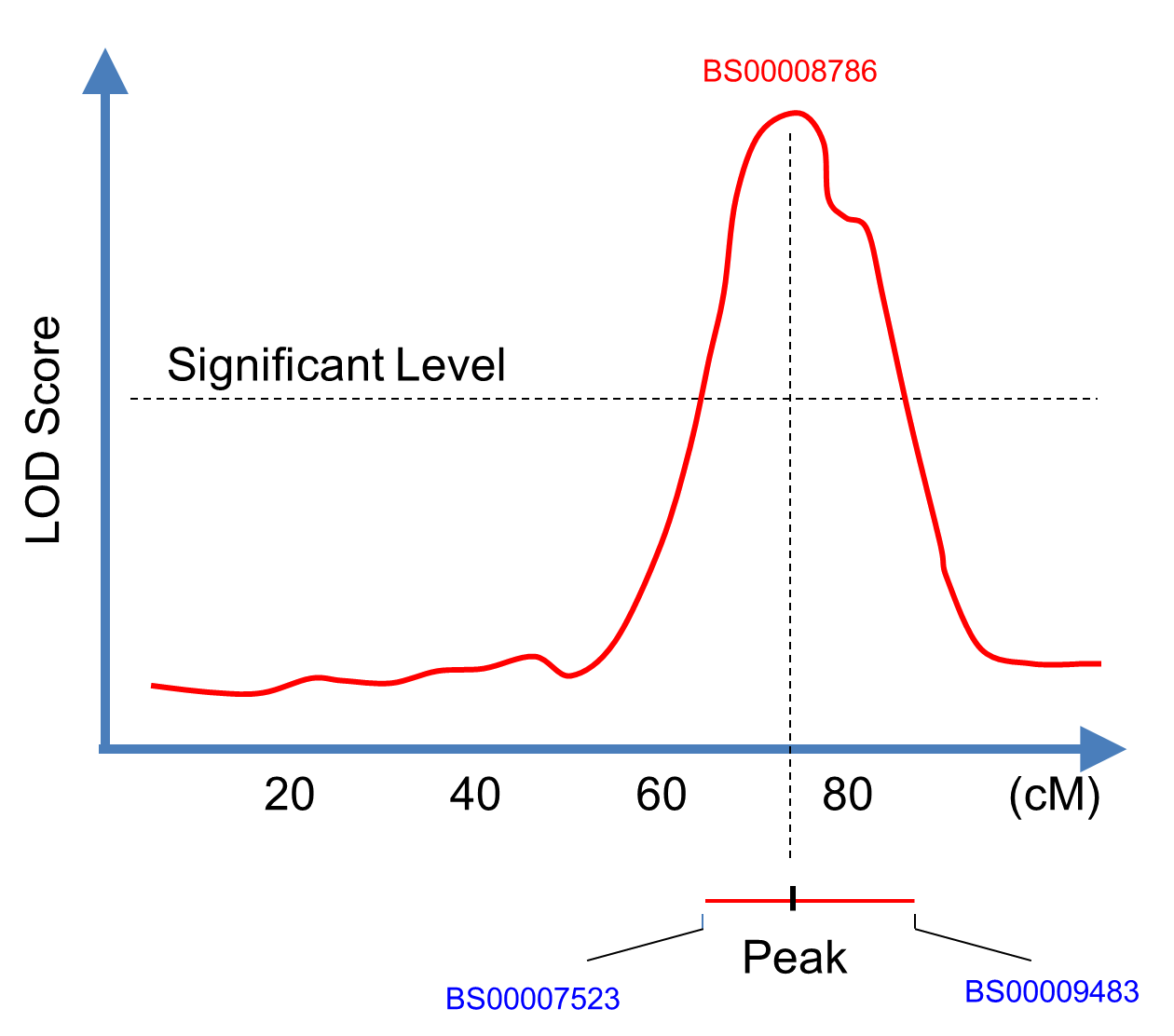 | QTL database - Select a phenotype to obtain all QTLs associated with it. |
 | Cross-species SNPs - Find SNP containing sequences that align to regions in Brachypodium, Rice and Sorghum. |
 | SNP Probe Locator - Search for SNP probes located on the same chromosome in the IWGSC v1.0 assembly. |
 | Dendrograms - View dendrograms based on data obtained using the 820K Array or 35K Array. |
 | Putative Introgression Plotter - Search for putative introgressions in wheat varieties. |
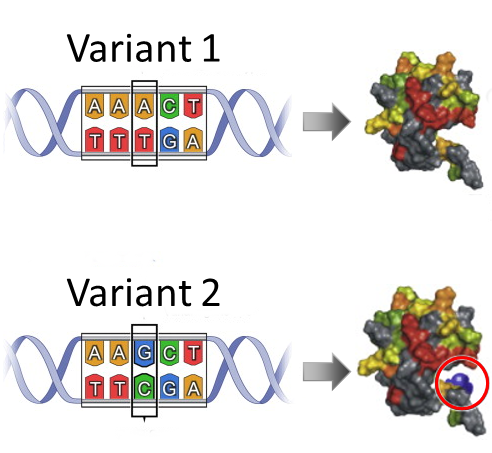 | Functional SNPs - Search non-synonymous (missense) SNP; i.e., SNPs that change protein amino acid sequence. |
 | Minimal Marker set - see the set of 32 SNP markers and identify varieties and their profiles. |

 -->
-->

 -->
-->