Making a request to the web services
There are a number of ways of requesting information from the CerealsDB web services. The simplest of these is to use cURL command line tool for getting or sending files using URL syntax. There is a download wizard for cURL if you do not have it installed on your system, and this can be accessed here.
Before using cURL you will need to create a file containing your JSON request, in this example we create a file called get_services_request.json and paste in the initial JSON request for the services that CerealsDB has available.
We can then send this file containing the JSON request to CerealsDB using the following cURL command:This should return a page of JSON code that we can parse using a script, or a request for data can be constructed and submitted by cURL. An example JSON response to extract a known contig (eg BC000000118) from CerealsDB is shown below. The contig services is set to true and we aren't requesting information from the Searchservice, so this has been set to false. In addition we aren't interested in SNP information, so the SNP_id "current_value" has been left empty.
The JSON code from above can be pasted into a new file - you could call it 'data_request.json' and this will be used in the next cURL request. From the initial JSON response we can see that the URL we need to direct this query is http://www.cerealsdb.uk.net/cerealgenomics/cgi-bin/parse_json_v3.cgi so the syntax would be:
You should receive a response that looks something like:
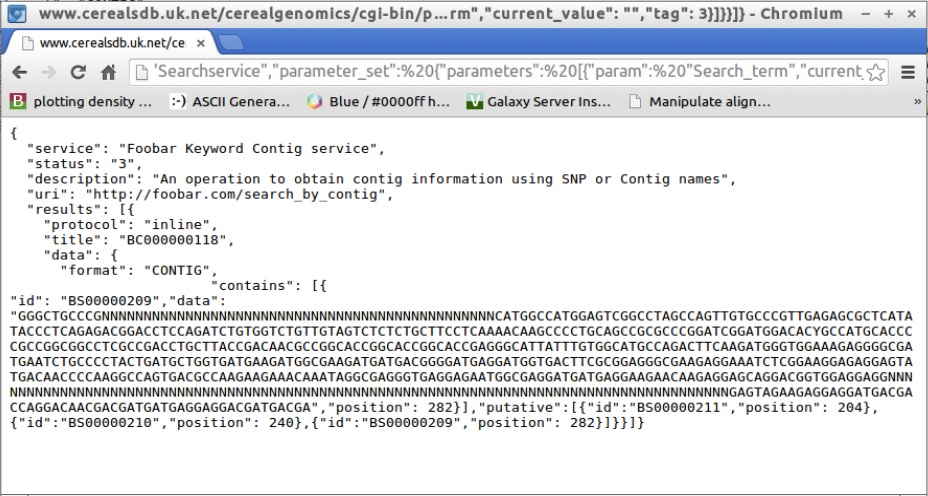
The JSON response describes four SNPs and the positions in the sequence that they are located in. The first SNP also includes a data key, the value of which is the nucleotide sequence of the contig that the SNPs are located in.
Of course, as we are using a RESTful web service, an alternative to using curl would be to paste the JSON query into a web browser's address bar. This would then display the JSON response from CerealsDB within your browser, and as JSON is designed to be human readable, it can help developers to quickly check that their constructed requests are working properly. To try this, simply copy and paste the text below into your browsers address bar:
This is the address to the CerealsDB JSON script:
http://www.cerealsdb.uk.net/cerealgenomics/cgi-bin/parse_json_v3.cgi?=with the JSON code above appended to the end of it. If it's working correctly, you should see the JSON code displayed in your browser like the example below.
Most users of the web services won't be using curl or a web browser to access the data in CerealsDB. They will be writing their own code in Perl or Python (or any language they want) to interrogate our database. There are example scripts on the "Sample templates" link in the webservices menu to the right of this page.
Based at the University of Bristol with support from BBSRC.

