WheatBP NEW
CerealsDB home WheatBP home FAQs Contact us
Wheat evolution Wheat development Wheat farming Milling Baking and brewing Wheat research SNPs SNP discovery
Our publications Related Web-sites
SNP discovery and genotyping in wheat
Food security is a global concern and substantial yield increases in cereal crops are required to feed the growing world population. Wheat is one of the three most important crops for human and livestock feed. However, the complexity ot its genome, coupled with a decline in genetic diversity within modern elite cultivars has hindered the application of marker-assisted selection (MAS) in breeding programs. A crucial step in the successful application of molecular breeding programs is the development of cheap, easy to use molecular markers, such as single nucleotide polymorphisms (SNPs). However, SNP discovery and genotyping in wheat is complicated by the relatively large numbers of polymorphisms seen in homoeologous and paralogous genes compared to the relatively infrequent varietal polymorphisms (Barker and Edwards, 2009). To overcome these challenges we have used a novel sequence alignment and assembly approach to mine SNPs from next generation sequence data and validated putative SNPs using a new high-throughput genotyping procedure that is capable of distinguishing inter-varietal from inter-genomic polymorphisms.
SNP discovery
Three complimentary approaches have been taken to identify putative varietal SNPs. First, screening of the publicly available wheat EST database (generated from numerous cDNA libraries, from a random collection of varieties relating to different stages of grain development and various stress treatments) yielded 3500 putative varietal SNPs in 8668 sequences. Secondly, normalised whole-seedling cDNA from five wheat varieties (Avalon, Cadenza, Rialto, Savannah, Recital) was sequenced on the Illumina GAIIx platform. For each line, we generated between 24 and 45 million, 75-base pair (bp) paired-end reads. SNP discovery (see Allen et al., 2011 for details), resulted in the identification of 14 078 putative SNPs in 6255 distinct reference sequences covering 2.7 megabases in total. On average, this equates to five varietal SNPs per kilobase in the reference sequences containing one or more SNP. The third approach used a Nimblegen capture array (Roche) with probe sequences designed using the Chinese Spring 5x genome sequence. The array was used to enrich genomic DNA samples for genic reads in seven UK wheat varieties (Alchemy, Avalon, Cadenza, Rialto, Hereward, Savannah, Xi19) enabling a greater depth of sequence coverage in coding regions. The enriched DNA samples were sequenced on the Illumina GAIIx platform and yielded between 24 and 45 million 75-base pair reads for each variety from which 95,267 novel putative varietal SNPs were mined.
SNP validation and characterisation
To date, over 2000 SNPs have been validated as polymorphic between different wheat varieties using the KASP genotyping platform (LGC Genomics - formerly KBioscience). The wheat varieties were selected following a survey of UK academics and wheat breeders to ensure that material of use to the whole community was represented; 21 hexaploid wheat varieties, a diploid and a tetraploid were included. Based upon the SNP genotyping data of these 23 varieties, the polymorphism information content (PIC) values of the validated SNPs varied from 0.080 to 0.375 with an average value of 0.300.
In wheat the majority (~90%) of the KASP probes detect both the polymorphic and non-polymorphic homoeologous loci. While such probes are suitable for screening inbred wheat varieties, screening heterozygous material is more problematical. To overcome this, we investigated the possibility of converting a standard KASP probe to a homoeologous-specific KASP probe by incorporating homoeologous SNPs in the reverse primer sequence (Figure 1). Use of the redesigned homoeologous-specific primers to screen an F2 population confirmed that they were capable of discriminating between homozygotes and heterozygotes (Figure 1e).
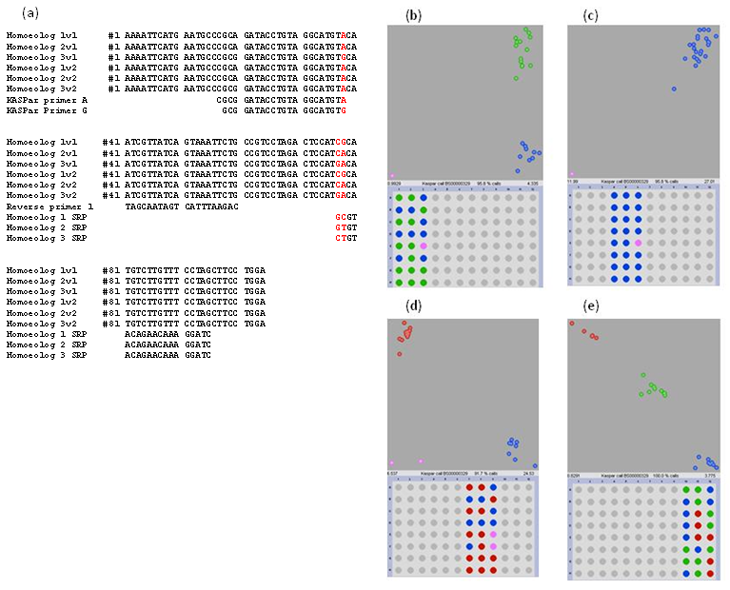
Figure 1. Effect of primer design of specificity of the KASP reaction. (a) Sequence of region around SNP BS00000329 in the three wheat homoeologs and across two varieties (v1 and v2) together with the six probes designed for this region. The varietal SNP (A/G) and the homoeologous SNPs are highlighted. The reverse primer 1 and the three homoeolog specific primers (SRP) are shown as designed in the reverse compliment to the BS00000329 sequence. (b-d) KASP plots following amplification with the various primer combinations. For b-c the standard 20 hexaploid varieties were used together with a negative control (E3, E6 and E9) and a diploid, tetraploid and resulting synthetic line. In all cases the KASP primers A and G were used. In (b) The generic reverse primer was used; in (c) Homoeolog 1 SPR was used whilst in (d) Homoeolog 3 SPR was used. No results are presented for Homeolog 2 SPR as this gave the same result as Homoeolog 1 SPR. (e) KASP plot of F2 derived material following amplification with the KASP primers A and G together with the Homoeolog 3 SRP primer. In (e) both homozygotes (blue and red spots) and heterozygotes (green spots) are shown as discrete clusters.
Genetic Map Construction
A linkage map is often the first step towards understanding genome assembly and evolution and provides an essential framework for mapping agronomic traits of interest. In order to place our SNP markers on a genetic map we scored 190 individuals from an Avalon x Cadenza doubled haploid population for 1208 loci using KASP genotyping. Using the information generated, in conjunction with the 574 non-SNP markers previously available for this population (provided by the Wheat Genetic Improvement Network), we were able to map 1165 SNP markers to 21 linkage groups representing chromosomes, with a further 43 loci mapping to unassigned linkage groups. The mapped sequences were BLAST searched against the Brachypodium distachyon genome sequence; where sufficient markers existed, as for wheat linkage group 2 versus B. distachyon chromosome 5, the relationship was consistent with known chromosomal relationships (International Brachypodium Initiative, 2010 and Allen et al., 2011).

SNP map of chromosome 1A.
Conclusion
This study represents an important step forward in wheat genetics as the first public linkage map for hexaploid wheat containing several hundred SNP markers. In addition, we believe that it is also the first reported use of KASP-based technology to both genotype wheat varieties and generate a linkage map. Using this and similar technology, we believe that it will be possible for wheat breeders to achieve one of their most important goals; to rapidly and cheaply genotype thousands of plants with a large and flexible number of markers. Our studies have also shown that there is need for further SNPs, especially for the D-genome and the homoeologous group 4 chromosomes, but with continued SNP development genome-wide association studies will become possible in wheat, in the near future. The extensive genomics resources developed by whole genome shotgun sequencing of Chinese Spring, coupled to re-sequencing multiple breeding lines, promises to dramatically increase the number of informative SNPs, permitting unprecedented levels of precision genetic analysis in wheat breeding.